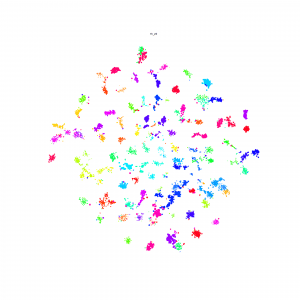
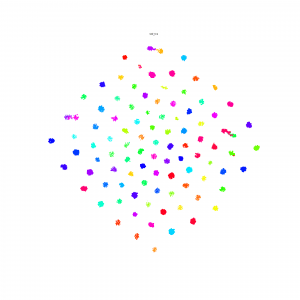
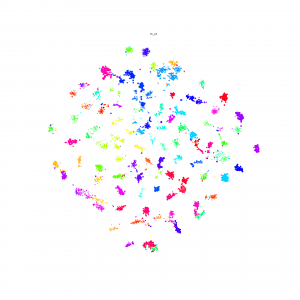I made Npair loss and Proxy loss embedding method available on the Lilou: /pless_nfs/home/share_projects/
Category: Uncategorized
Using NCSA Server to Find the Crop Position of Raw Data
Leaf length/width pipeline
The leaf length/width pipeline for season 6 is running on DDPSC server. This is going to be finished in next week.
The pipeline currently running finds the leaves first instead of plots. So I rewrote the merging to fit this method.
Leaf Curvature
I'm digging into the PCL (Point Cloud Library) to see if we could apply this library to our point cloud data. This library is originally developed on C++. There is an official python binding project under development. But there are not too many activities on that repo for years. (Also there is a API for calculate the curvature is not implemented on this binding.) So should we working on some point cloud problems on C++? If we are going to keep working on the ply data, considering the processing speed for point cloud and the library, this seems like a appropriate and viable way to work with.
Or, at least for the curvature, I could implement the method used in PCL with python. Since we already have the xyz-map. Finding the neiberhood could be faster than on the ply file. Then the curvature could be calculated with some differential geometry methods
PCL curvature: http://docs.pointclouds.org/trunk/group__features.html
Python bindings to the PCL: https://github.com/strawlab/python-pcl
Reverse Phenotyping
Since the ply data are too large (~20 TB) to download(~6 MB/s). I created a new pipeline to find only the cropping position with ply file. So that I can run this on NCSA server and use those information to crop the raw data on our server. This is running on NCSA server now and I'm working on the cropping procedure.
I'm going to try Triplet loss, Hong's NN loss and Magnet loss to train the new data and do what we did before to visualize the result.
Yoking Many t-SNEs
This post has several purposes.
FIRST: We need a better name or acronym than yoked t-sne. it kinda sucks.
- Loosely Aligned t-Sne
- Comparable t-SNE (Ct-SNE)?
- t-SNEEZE? t-SNEs
SECOND: How can we "t-SNEEZE" many datasets at the same time?
Suppose you are doing image embedding, and you start from imagenet, then from epoch to epoch you learn a better embedding. It might be interesting to see the evolution of where the points are mapped. To do this you'd like to yoke (or align, or tie together, or t-SNEEZE) all the t-SNEs together so that they are comparable.
t-SNE is an approach to map high dimensional points to low dimensional points. Basically, it computes the similarity between points in high dimension, using the notation:
P(i,j) is (something like) how similar point i is to point j in high dimensions --- (this is measured from the data), and
Q(i,j) is (something like) how similar point i is to point j in low dimension.
the Q(i,j) is defined based on where the 2-D points are mapped in the t-SNE plot, and the optimization finds 2-D points that makes Q and P as similar as possible. Those points might be defined as (x(i), y(i)).
With "Yoked" t-SNE we have two versions of where the points go in high-dimesional space, so we have two sets of similarities. So there is a P1(i,j) and a P2(i,j)
yoked t-SNE solves for points x1, y1 and x2,y2 so that the
- Q1 defined by the x1, y1 points is similar to P1, and the
- Q2 defined by the x2,y2 points are similar to P2 and the
- x1,y1 points are similar to x2,y2
by adding this last cost (weight by something) to the optimization. If we have *many* high dimensional points sets (e.g. P1, P2, ... P7, for perhaps large versions of "7") what can we do?
Idea 1: exactly implement the above approach, with steps 1...7 talking about how each embedding should have Q similar to P, and have step 8 penalize all pairwise distances between the x,y embeddings for each point.
Idea 2: (my favorite?). The idea of t-SNE is to find which points are similar in high-dimensions and embed those close by. I wonder if we can find all pairs of points that are similar in *any* embedding. So, from P1... P7, make Pmax, so that Pmax(i,j) is the most i,j are similar in any high-dimensional space. Then solve for each other embedding so that it has to pay a penalty to be different from Pmax? [I think this is not quite the correct idea yet, but something like this feels right. Is "Pmin" the thing we should use?]
Optimization to simultaneously solve for light & camera locations
Earlier this week, Dr. Pless, Abby and I worked through some of the derivations of the ellipse equations in order to better understand where our understanding of them may have gone awry. In doing so, I now believe there is a problem with the fact that we are not considering the direction of the surface normals of the glitter. It seems to me that this is the reason for the need for centroids to be scattered around the light/camera - this is how the direction of surface normals is seen by the equations; however, this is not realistically how our system is set up (we tend to have centroids all on one side of the light/camera).
I did the test of generating an ellipse, finding points on that ellipse, and then using my method of solving the linear equations to try and re-create the ellipse using just the points on the ellipse and their surface normals (which I know because I can find the gradient at any point on the ellipse, and the surface normal is the negative of the gradient).
Here are some examples of success when I generate an ellipse, find points on that ellipse (as well as their surface normals), and then try to re-create the ellipse:




Funny story - the issue of using all points on one side of the ellipse and it not working doesn't seem to be an issue anymore. Not sure whether this is a good thing or not yet...look for updates in future posts about this!
We decided to try and tackle the optimization approach and try to get that up and running for now, both in 2D and 3D. I am optimizing for both the light and the camera locations simultaneously.
Error Function
In each iteration of the optimization, I am computing the surface normals of each centroid using the current values of the light and camera locations. I then want to maximize the sum of the dot products of the actual surface normals and the calculated surface normals at each iteration. Since I am using a minimization function, I want to minimize the negative of the sum of dot products.
error = -1*sum(dot(GT_surf_norms, calc_surf_norms, 2));
2D Results
Initial Locations: [15, 1, 15, 3] (light_x, light_y, camera_x, camera_y)
I chose this initialization because I wanted to ensure that both the light & camera start on the correct side of the glitter, and are generally one above the other, since I know that this is their general orientation with respect to the glitter.
Final Locations: [25, 30, 25, 10]
Actual Locations: [25, 10, 25, 30]
Final Error: 10
So, the camera and light got flipped here, which sensibly could happen because there is nothing in the error function to ensure that they don't get flipped like that.
3D Results
Coming soon!
Other things I am thinking about in the coming days/week
- I am not completely abandoning the beautiful ellipse equations that I have been working with. I am going to take some time to analyze the linear equations. One thing I will try to understand is whether there is variation in just 1 axis (what we want) or more than 1 axis (which would tell us that there is ambiguity in the data I am using when trying to define an ellipse).
- After I finish writing up the optimization simulations in both 2D and 3D, I will also try to analyze the effect that some noise in the surface normals may have on the results of the optimization.
Nearest Neighbor Loss for metric learning
The problem of common metric loss(triplet loss, contrast divergence and N-pair loss)
For a group of images belong to a same class, these loss functions will randomly sample 2 images from the group and force their dot-product to be large/euclidean distance to be small. And finally, their embedding points will be clustered in to circle shape. But in fact this circle shape is a really strong constraints to the embedding. There should be a more flexibility for them to be clustered such as star shape and strip shape or a group image in several clusters.
Nearest Neighbor Loss
I design a new loss to loss the 'circle cluster' constraints. First, using the idea in the end of N-pair loss: constructing a batch of images with N class and each class contain M images.
1 finding the same label pair
When calculating the similarity for image i, label c, in the batch, I first find its nearest neighbor image with label c as the same label pair.
2.set the threshold for the same pair.
if the nearest neighbor image similarity is small than a threshold, then I don't construct the pair.
3.set the threshold for the diff pair.
With the same threshold, if the diff label image have similarity smaller than the threshold than it will not be push away.
T-SNE result
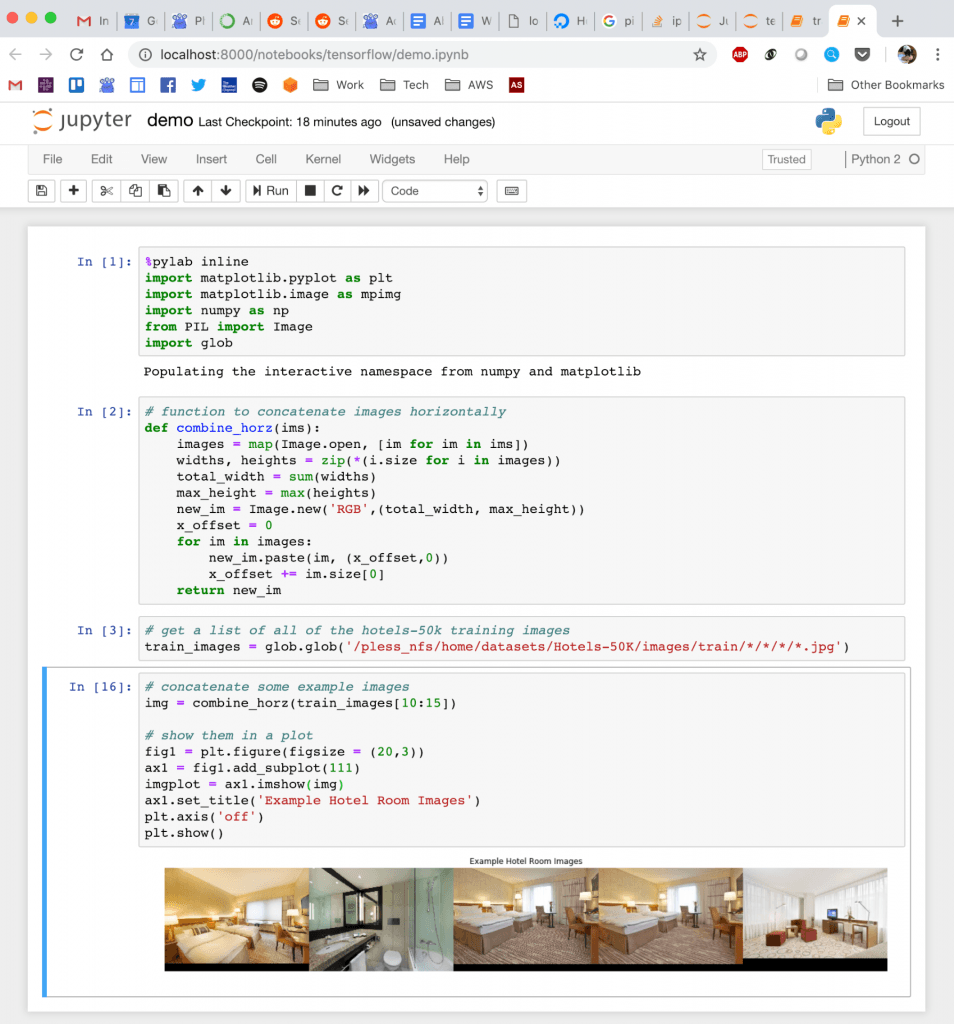
Accessing Server Jupyter Notebooks on Your Local Machine
For the last few years, I been doing development by writing code, deploying it to the server, running stuff on the server without any GUI, and then downloading anything that I want to visualize. This has been a huge pain! We work with images and having that many steps between code debugging and visualizing things is stupidly inefficient, but sorting out a better way of doing things just hadn't boiled up to the top of my priority list. But I've been crazy jealous of the awesome things Hong has been able to do with jupyter notebooks that run on the server, but which he can operate through the browser on his local machine. So I asked him for a bit of help getting up and running so that I could work on lilou from my laptop and it turns out it's crazy easy to get up and running!
I figured I would detail the (short) set of steps in case other folk's would benefit from this -- although maybe all you cool kids already know about running iPython and think I'm nuts for having been working entirely without a GUI for the last few years... 🙂
On the server:
- Install anaconda:
Get the link for the appropriate anaconda version
On the server run wget [link to the download]
sh downloaded_file.sh
- Install jupyter notebook:
pip install —user jupyter
Note: I first had to run pip install —user ipython (there was a python version conflict that I had to resolve before I could install jupyter)
- Generate a jupyter notebook config file:jupyter notebook --generate-config
- In the python terminal, run:
from notebook.auth import passwd
passwd()
This will prompt you to enter a password for the notebook, and then output the sha1 hashed version of the password. Copy this down somewhere.
- Edit the config file (~/.jupyter/jupyter_notebook_config.py):
Paste the sha1 hashed password into line 276:
c.NotebookApp.password = u'sha1:xxxxxxxxxxxxxx'
- Run “jupyter notebook” to start serving the notebook
Then to access this notebook locally:
- Open up the ssh tunnel:ssh -L 8000:localhost:8888 username@lilou.seas.gwu.edu
- In your local browser, go to localhost:8000
- Enter the password you created for your notebook on the server in step 4 above
- Create iPython notebooks and start running stuff!
Quick demo of what this looks like:
Fighting hard on pipelines
Leaf length + width
Processing speed
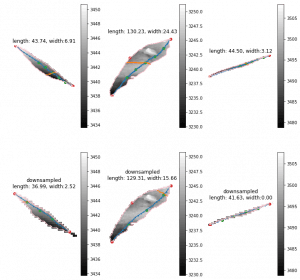
Currently we have more than 20000 scans for one season. Two scanners are included in the scanner box. Then we have 40000 strips for one season. Previously, 20000 stripes leaf length last 2 weeks. The new leaf length + width costs about double. Then we are going to spend about 2 month. This is totally unacceptable. I'm finding the reason and trying to optimize that. The clue that I have is the large leaves. The 2x length will cause 4x points in the image. Then there going to have 2x points on edge. The time consuming of longest shortest path among edge points will be 8-16x. I've tried downsample the leaf with several leaves. The difference of leaves length is about 10-15 mm. But the leaf width seems more unreasonable. It should relevant to the how far should be considered as neighborhood of points on the graph. I'm trying to downsample the image to a same range for both large and small leaves to see if this could be solved.
Pipeline behavior
The pipeline was created to run locally on lab server with per plot cropping. However, we are going to using this pipeline on different server, with different strategies. also this is going to be used as a extractor. So I modified the whole pipeline structures to make it more flexible and expandable. Features like using different png and ply data folder, specify the output folder ,using start and end date to select specific seasons, download from Globus or not and etc are implemented.
Reverse Phenotyping
Data
Since Zongyang suggested me cropping the plots from scans by point cloud data. I'm working on using point cloud to regenerate the training data to revise the reverse phenotyping. Since the training data we used before may have some mislabeling. I'm waiting for the downloading since it's going to cost about 20 days. Could we have a faster way to get data?
Compilation of Visualizations
In this post, I am going to run through everything that I have tried thus far, and show some pictures of the results that each attempt has rendered. I will first discuss the results of the experiments involving a set of vertically co-linear glitter, 10 centroids. Then, I will discuss the results of the experiments involving 5 pieces of glitter placed in a circle around the camera & light, such that the camera & light are vertically co-linear.
Calculation of Coefficients
In order to account for the surface normals having varying magnitudes (information that is necessary in determining which ellipse a piece of glitter lies on), I use the ratios of the surface normal's components and the ratios of the gradient's components (see previous post for derivation).
Once I have constructed the matrix A as follows:
A = [-2*SN_y * x, SN_x *x - SN_y *y, 2*SN_x *y, -SN_y, SN_x], derived by expanding the equality of ratios and putting the equation in matrix form. So, we are solving the equation: Ax = 0, where x = [a, b, c, d, e]. In order to solve such a homogeneous equation, I am simply finding the null space of A, and then using some linear combination of these vectors to get my coefficients:
Z = null(A);
temp = ones(size(Z,2),1);
C = Z*temp
Using these values of coefficients, I can then calculate the value of f in the implicit equation by directly solving for it for each centroid.
1. Glitter is vertically co-linear
For the first simulation, I placed the camera and light to the right of the glitter line, and vertically co-linear to each other, as seen in the figure to the left. Here, the red vectors are the surface normals of the glitter and the blue vectors are the tangents, or the gradients, as calculated based on the surface normals.
- Using the first 5 pieces of glitter:
- Coefficients are as follows:
- a = -0.0333
- b = 0
- c = 0
- d = 0.9994
- e = 0
- No plot - all terms with a y are zeroed out, so there is nothing to plot. Clearly not right...
- Coefficients are as follows:
- Using the last 5 pieces of glitter:
- Same results as above.
This leads me to believe there is something unsuitable about the glitter being co-linear like this.
 For the second simulation, I placed the camera and the light to the right of the glitter line, but here they are not vertically co-linear with each other, as you can see in the figure to the left.
For the second simulation, I placed the camera and the light to the right of the glitter line, but here they are not vertically co-linear with each other, as you can see in the figure to the left.
- Using the first 5 pieces of glitter:
- Coefficients are as follows:
- a = 0.0333
- b = 0
- c = 0
- d = -0.9994
- e = 0
- No plot - all terms with a y are zeroed out, so there is nothing to plot. Clearly not right...
- Coefficients are as follows:
- Using the last 5 pieces of glitter:
- Same results as above.
If we move the camera and light to the other side of the glitter, there is no change. Still same results as above.
2. Glitter is not vertically co-linear
 In this experiment, the glitter is "scattered" around the light and camera as seen in the figure to the left.
In this experiment, the glitter is "scattered" around the light and camera as seen in the figure to the left.
I had a slight victory here - I actually got concentric ellipses in this experiment when I move one of the centroids so that it was not co-linear with any of the others:
In the process of writing this post and running through all my previous failures, I found something that works; so, I am going to leave this post here. I am now working through different scenarios of this experiment and trying to understand how the linearity of the centroids affects the results (there is definitely something telling in the linearity and the number of centroids that are co-linear with each other). I will try to have another post up in the near future with more insight into this!
The variance of embedding space
In the image embedding task, we always focus on the design of the loss and make a little attention on the output/embedding space because the high dimensional space is hard to image and visualized. So I find an old tool can help us understand what happened in our high-dimension embedding space--SVD and PCA.
SVD and PCA
SVD:
Given a matrix A size (m by n), we can write it into the form:
A = U E V
where A is a m by n matrix, U is a m by m matrix, E is a m by n matrix and V is a n by n matrix.
PCA
What PCA did differently is to pre-process the data with extracting the mean of data.
Especially, V is the high-dimensional rotation matrix to map the embedding data into a in coordinates and E is the variance of each new coordinates
Experiments
The feature vector is coming from car dataset(train set) trained with standard N-pair loss and l2 normalization
For a set of train set points after training, I apply PCA on with the points and get high-dimensional rotation matrix, V.
Then I use V to transform the train points so that I get the new representation of the embedding feature vectors.
Effect of apply V to the embedding points:
- Do not change the neighbor relationship
- ‘Sorting’ the dimension with the variance/singular value
Then let go back to view the new feature vectors. The first digit of the feature vectors represents the largest variance/singular value projection of V. The last digit of the feature vector represents the smallest variance/singular value projection of V
I scatter the first and the last digit value of the train set feature vectors and get the following plots. The x-axis is the class id and the y-axis is each points value in a given digit.
The largest variance/singular value projection dimension
The smallest variance/singular value projection dimension

We can see the smallest variance/singular value projection or say the last digit of the feature vector has very small values distribution and clustered around zero.
When comparing a pair of this kind of feature vector, the last digit contributes very small dot product the whole dot product(for example, 0.1 * 0.05 =0.005 in the last digit). So we can neglect this kind of useless dimension since it looks like a null space.
Same test with various embedding size
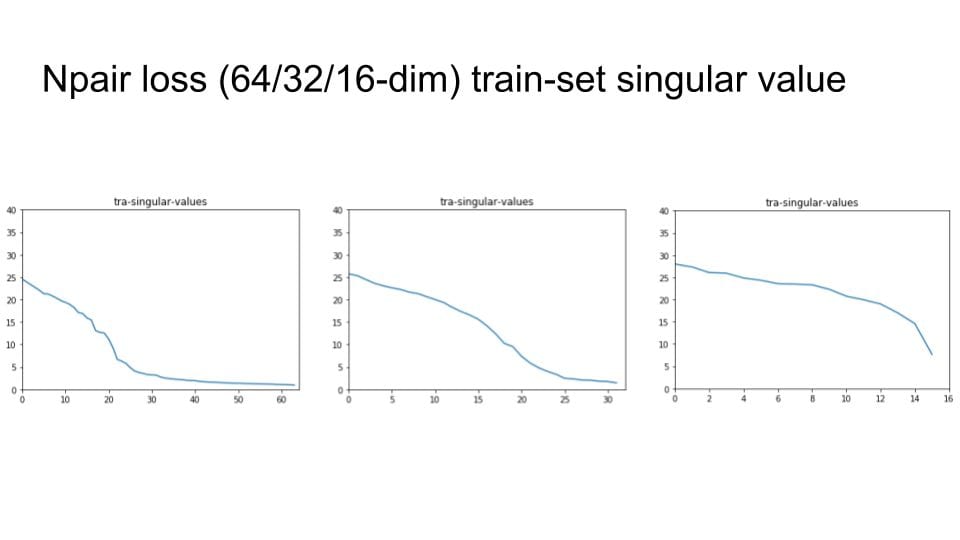
I try to change the embedding size with 64, 32 and 16. Then check the singular value distribution.
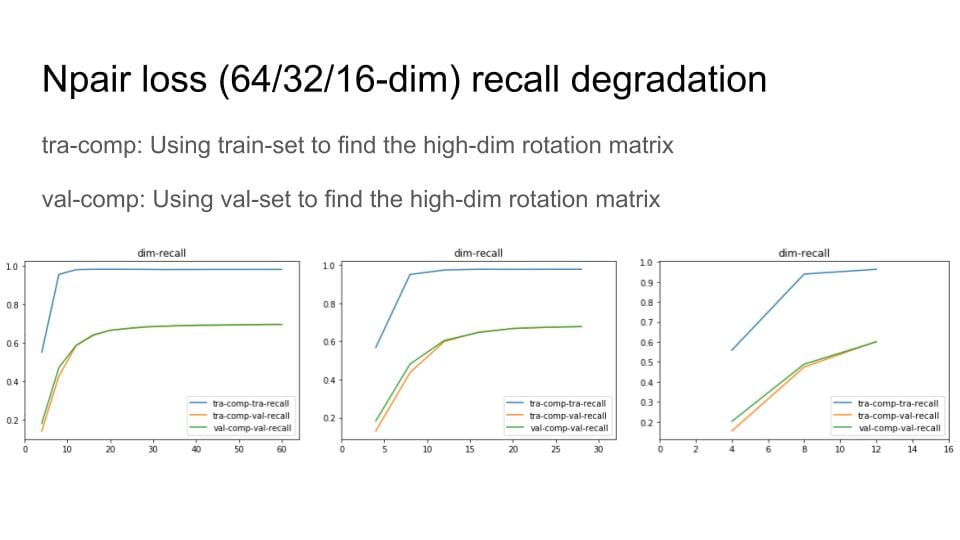
Then, I remove the digit with small variance and Do Recall@1 test to explore the degradation of the recall performance
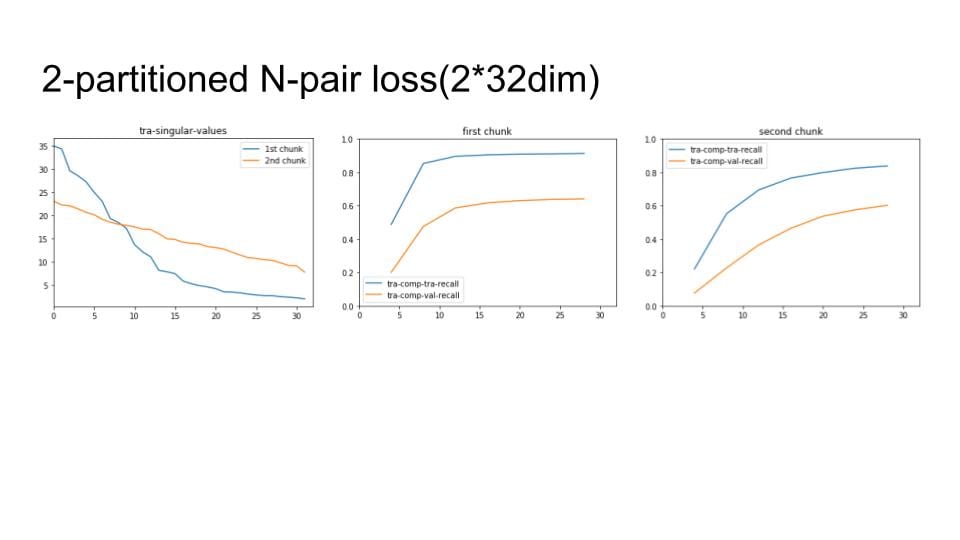
Lastly, I apply the above process to our chunks method
Photogeometry Research Group
Welcome to our lab blog!
We build cameras, algorithms, and systems that analyze images to understand our world in new ways. This page helps keep our lab organized:
Lab Members
Science and engineering thrives on diverse perspectives and approaches. Everyone is welcome in our lab, regardless of race, nationality, religion, gender, sexual orientation, age, or disabilities. Scientists from underrepresented groups are especially encouraged to join us.