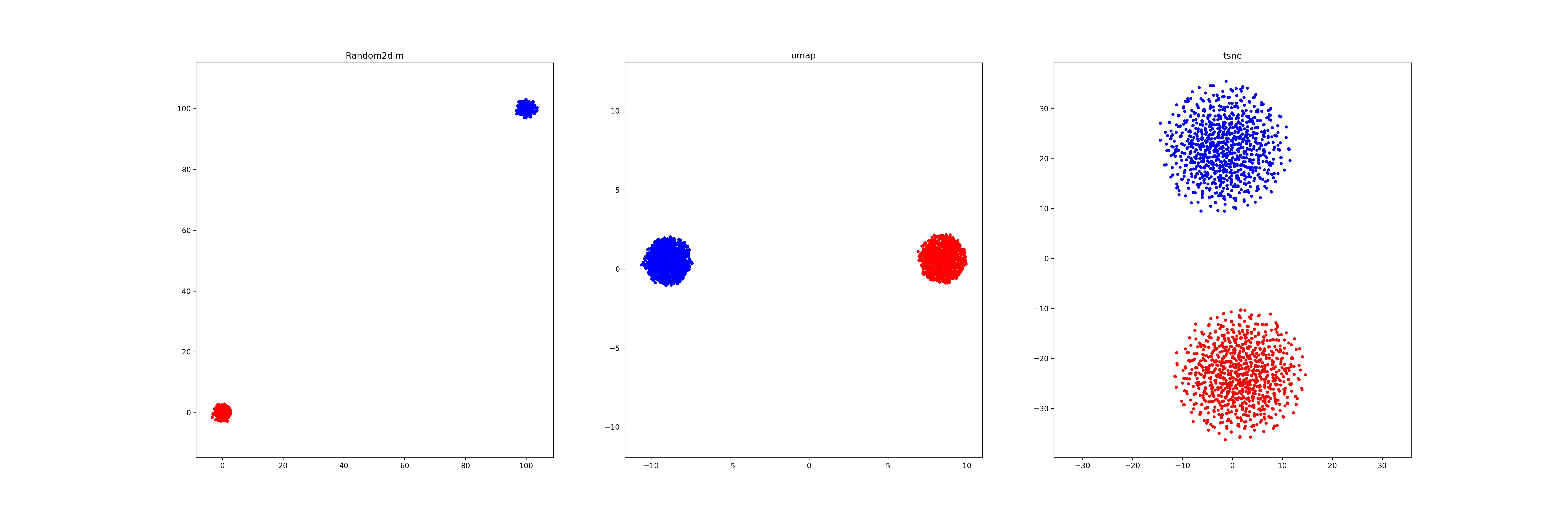
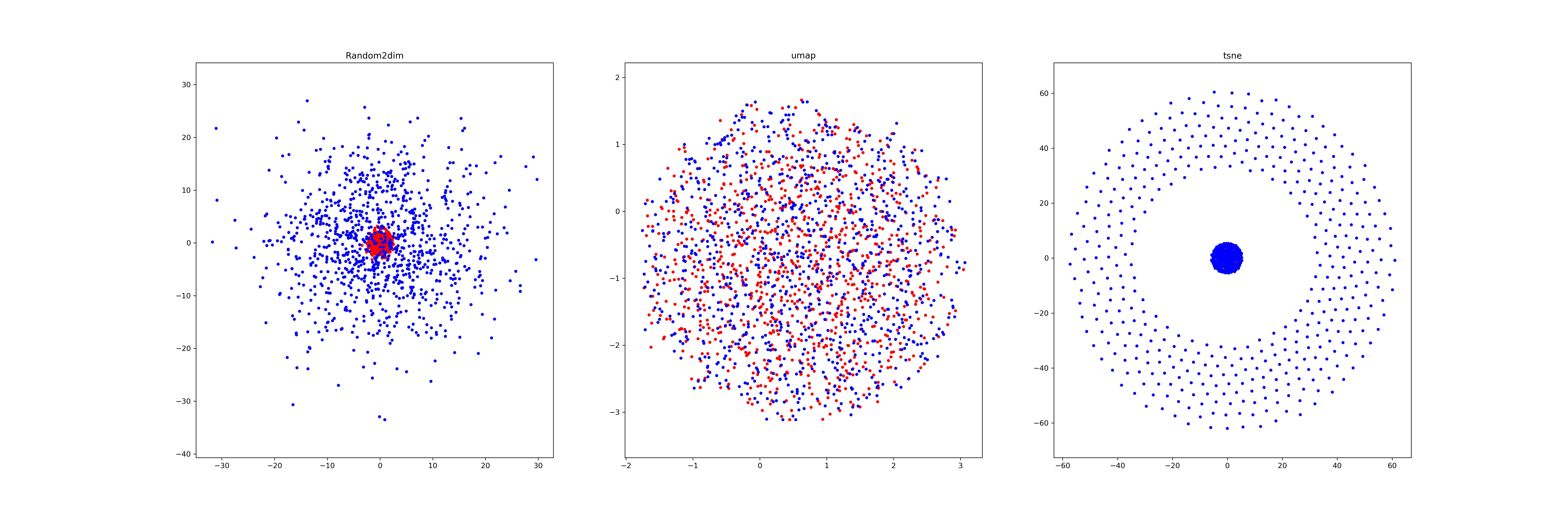
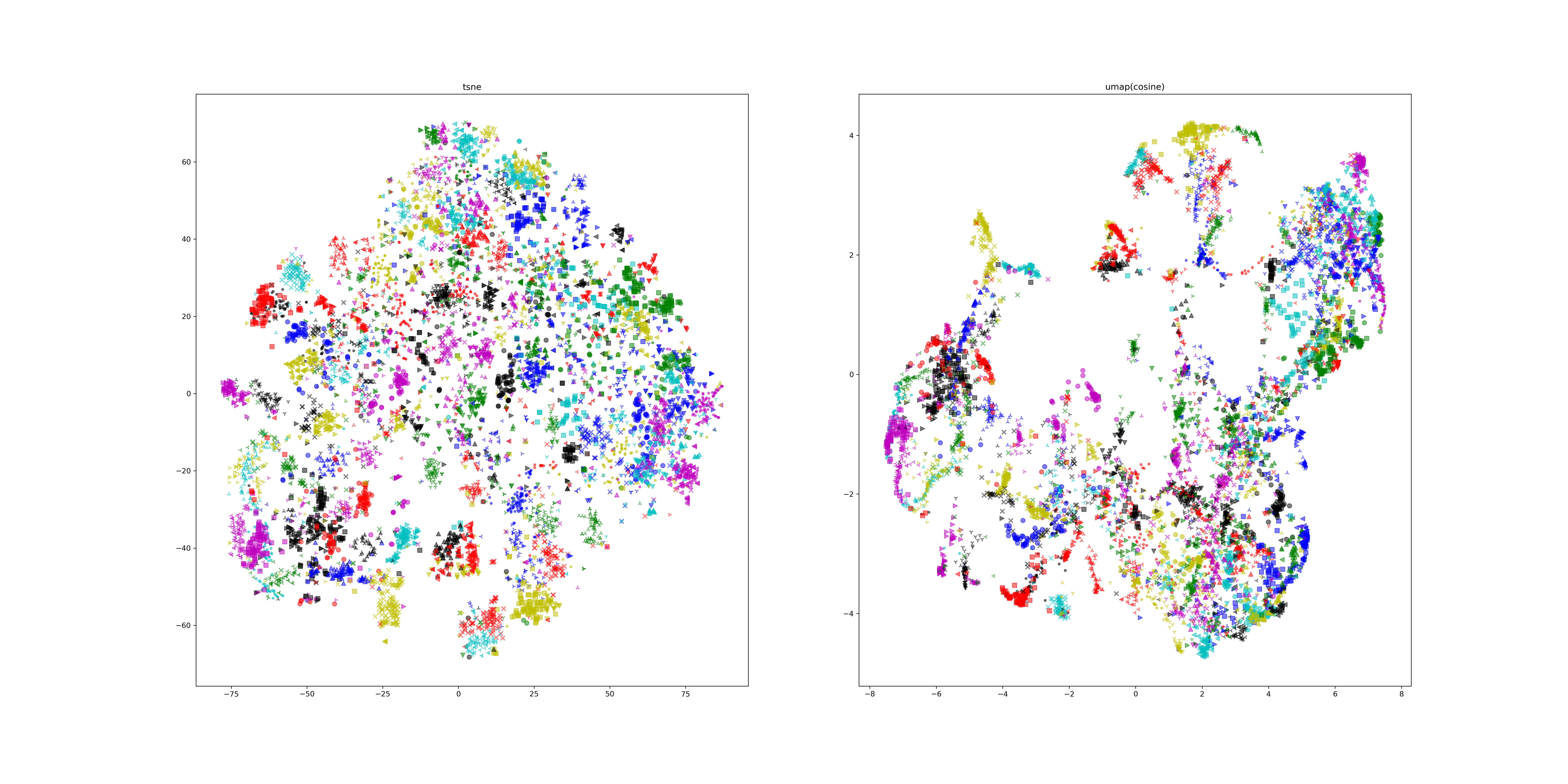
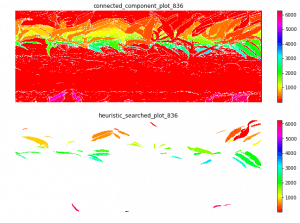
Last week, I find a problem of UMAP which is that if the high graph of a embedding representation is not connected, like NPair result on CAR training dataset, the optimizer of UMAP will keep push each cluster far away, which doesn't matter in visualization, but in TUMAP, we need to measure the loss of each map.
So, we try some different way to avoid or solve this problem.
First, we compute KL distance of normal UMAP with TUMAP result instead of compare their loss.
Second, we try to optimize the repulsive gradient of edge in the high graph instead of each connection of every two points. But the result of this method gonna wired.
And, I try add a 0 vector into the high dimensional vectors, and make it equally very far from each points when constructing high-D graph. It doesn't work.