Blog Post:
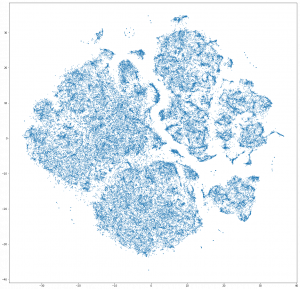
I’ve done lots of work on “embedding” over the last 20 years. In the early 2000’s this was called “manifold learning” and the goal was to do something like t-SNE:
Given a collection of images (or other high-D points) --- that you *think* have some underlying structure or relationship that you can define with a few parameters --- map those images into a low-dimensional space that highlights this structure.
Some of my papers in this domain are here.
https://www2.seas.gwu.edu/~pless/publications.php?search=isomap
This includes papers that promote the use of this for temporal super-resolution, using that structure to help segmentation of biomedical imagery, and modifications to these algorithms if the low-dimensional structure had cyclic structure (for example, if the images are of an object that has rotated all the way around, you want to map it to a circle, not a line).
Algorithms like Isomap, LLE, t-SNE and UMAP all follow this model. They take as input a set of images and map those images to a 2 or 3 dimensional space where the structure of the points is, hopefully, apparent and useful. These algorithms are interesting, but they *don’t* provide a mapping from any image into a low-dimensional space, they all just map the specific images you give them into a low dimensional space. It is often awkward to map new images to the low dimensional space (what you do is find the closest original image or two close original images and use their mapping and hope for the best).
These algorithms also assume that for very nearby points, simple ways of comparing points are effective (for example, just summing the difference of pixel values), and they try to understand the overall structure or relationship between points based on these trusted small distances. They are often able to do clustering, but they don’t expect to have any labelled input points.
What I learned from working in this space is the following:
- If you have enough datapoints, you can trust small distances. (Yes, this is part of the basic assumption of these algorithms, but most people don’t really internalize how important it is).
- You *can’t possibly* have enough datapoints if the underlying structure has more than a few causes of variation (because you kinda need to have an example of how the images vary due to each cause and you get to an exponential number of images you’d need very quickly).
- You can make a lot of progress if you understand *why* you are making the low-dimensional embedding, by tuning your algorithm to the application domain.
What does this tell us about our current embedding work? The current definition of embedding network has a different goal: Learn a Deep Learning network f that maps images onto points in an embedding space so that:
|f(a) - f(b)| is small if a,b are from the same category
and
|f(a) - f(b)| is large of a,b are from different categories.
This differs from the previous world in two fundamental ways:
- We are given labels that help tell us where our points should map to (or at least constraints on where they map), and
- We care about f(c) ... where do new points map?
- Usually, we don’t expect the original points to be close (in terms of pixel differences or other *easy* distance functions in the high dimensional space.
So we get extra information (in the form of labels of which different images should be mapped close to each other), at the cost of our input images maybe not being so close in their original space, and requiring that the mapping work for new and different images.
Shit, that’s a bad trade.
So we don’t have enough training images to be able to trace out clean subsets of similar images based on follow super similar images (you’d have to sample cars of many colors, in front of all backgrounds, with all poses, and all collections of which doors and trunks are open)
So what can you do?
Our approach\ (Easy-Positive) is something like “hope for the best”. We force the mapping to push the closest images of any category towards each other, and don’t ask the mapping to do more than that. Hopefully, this allows the mapping to find ways to push the “kinda close” images together. Hopefully, new data from new classes are also mapped close together.
What are other approaches? Here is a just-published to ArXiv paper that tries something: https://arxiv.org/pdf/1904.03436.pdf. They say, perhaps there are data augmentation steps (or ways of changing the images) that you *really* don’t believe should change your output vector. These might include slight changes in image scale, or flipping the image left/right, or slight skews of the image.
You could take an *enormous* number of random images, and say “any two images that are a slight modification of each other” should have exactly the same feature vector, and “any two images that are different should have different feature vectors.
This could be *added* to the regular set of images and triplet loss used to train a network and it would help force that network to ignore the changes that are in your data augmentation set. If you care most about geometric transformations, and really like formal math, you could read the following (https://arxiv.org/pdf/1904.00993.pdf).
The other thing we can, or tool that we can use to try to dig deeper into the problem domain, is ask more "scientific" questions. Some of our recent tools really help with this.
Question: When we “generalize” to new categories, does the network focus on the right part of the image?
How to answer this:
- Train a network on the training set. What parts of the image are most important for the test set?
- Train a network on the TEST set. What parts of the image are most important for the test set?
- We can do this and automatically compare the results.
- If the SAME parts of the image are important, then the network is generalizing in the sense that the features are computed on the correct part of the image.
This might be called “attentional generalization”.
Questions: If the network does focus does focus on the right part of the image, does it represent objects in a generalizable way?
How to answer this:
Something similar to the above, with a focus on image regions instead of the activation across the whole image.
This we might call “representational generalization" or "semantic generalization”