Leaf length + width
Processing speed
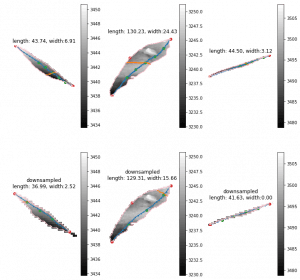
Currently we have more than 20000 scans for one season. Two scanners are included in the scanner box. Then we have 40000 strips for one season. Previously, 20000 stripes leaf length last 2 weeks. The new leaf length + width costs about double. Then we are going to spend about 2 month. This is totally unacceptable. I'm finding the reason and trying to optimize that. The clue that I have is the large leaves. The 2x length will cause 4x points in the image. Then there going to have 2x points on edge. The time consuming of longest shortest path among edge points will be 8-16x. I've tried downsample the leaf with several leaves. The difference of leaves length is about 10-15 mm. But the leaf width seems more unreasonable. It should relevant to the how far should be considered as neighborhood of points on the graph. I'm trying to downsample the image to a same range for both large and small leaves to see if this could be solved.
Pipeline behavior
The pipeline was created to run locally on lab server with per plot cropping. However, we are going to using this pipeline on different server, with different strategies. also this is going to be used as a extractor. So I modified the whole pipeline structures to make it more flexible and expandable. Features like using different png and ply data folder, specify the output folder ,using start and end date to select specific seasons, download from Globus or not and etc are implemented.
Reverse Phenotyping
Data
Since Zongyang suggested me cropping the plots from scans by point cloud data. I'm working on using point cloud to regenerate the training data to revise the reverse phenotyping. Since the training data we used before may have some mislabeling. I'm waiting for the downloading since it's going to cost about 20 days. Could we have a faster way to get data?