UMAP paper: https://arxiv.org/abs/1802.03426
Here are some attempt based on a python module, umap-learn
First, we try UMAP on some Gaussian dataset.
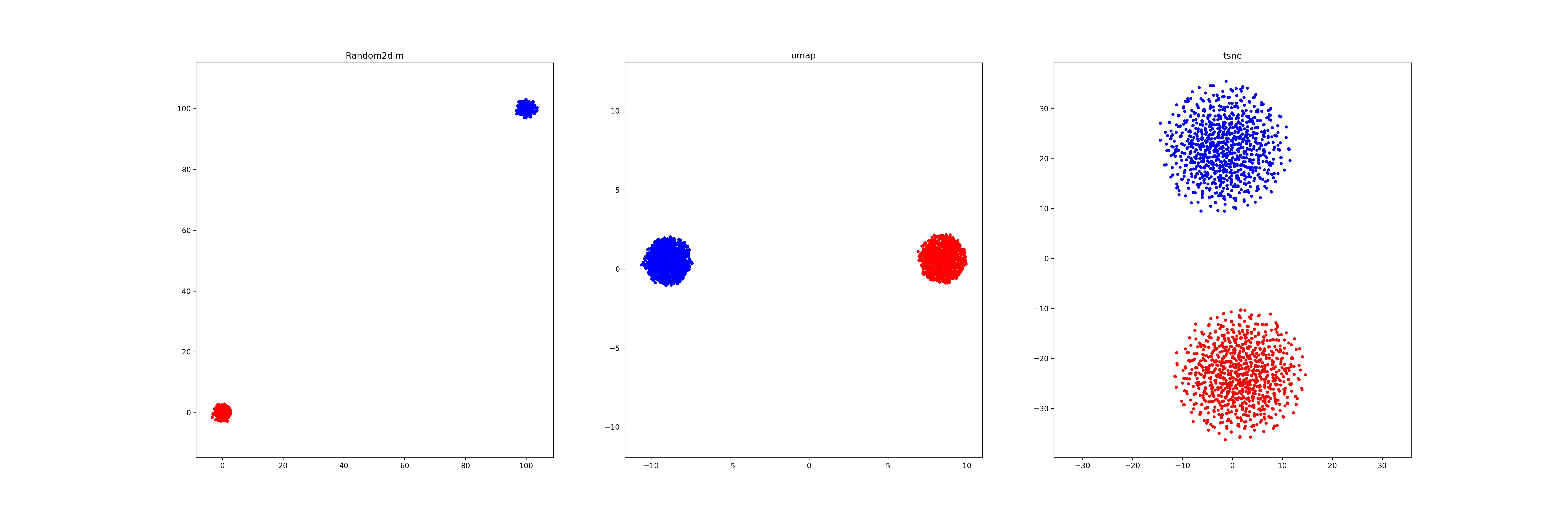
(1): We generate two different Gaussian distribution dataset (1000*64,1000*64) in different location (mean), and visualize it by a)random pick two dimension b) umap, c) tsne.
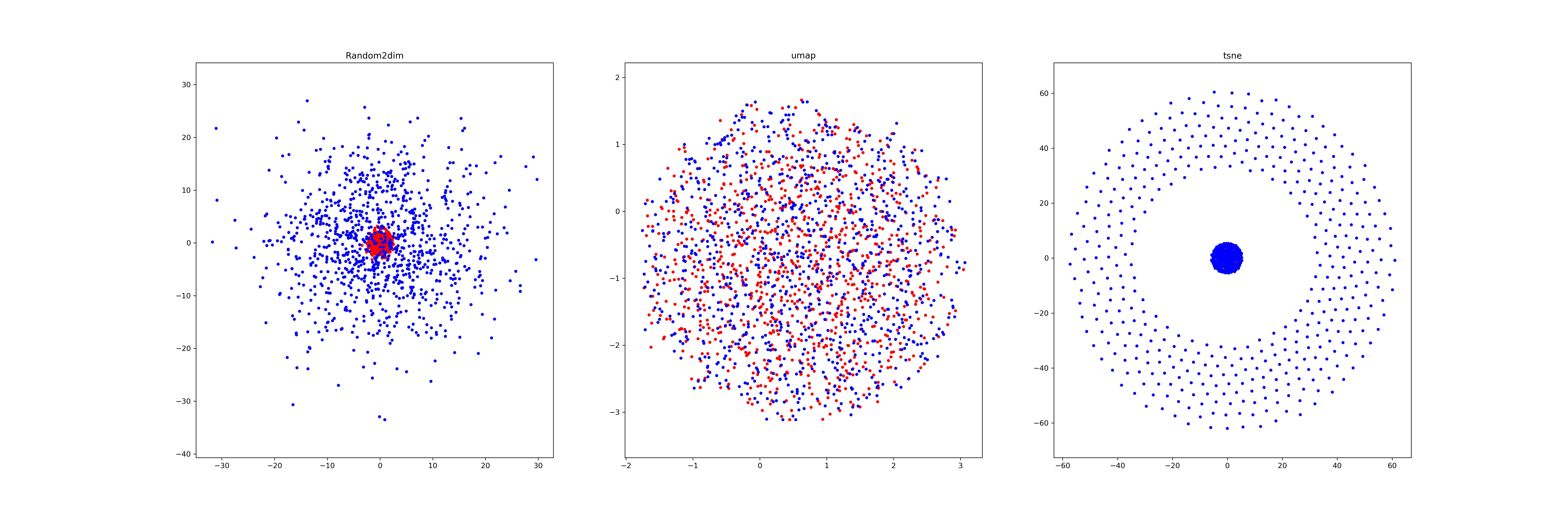
(2): We generate two different Gaussian distribution dataset (1000*64,1000*64) in different scale (std), and visualize it by a)random pick two dimension b) umap, c) tsne.
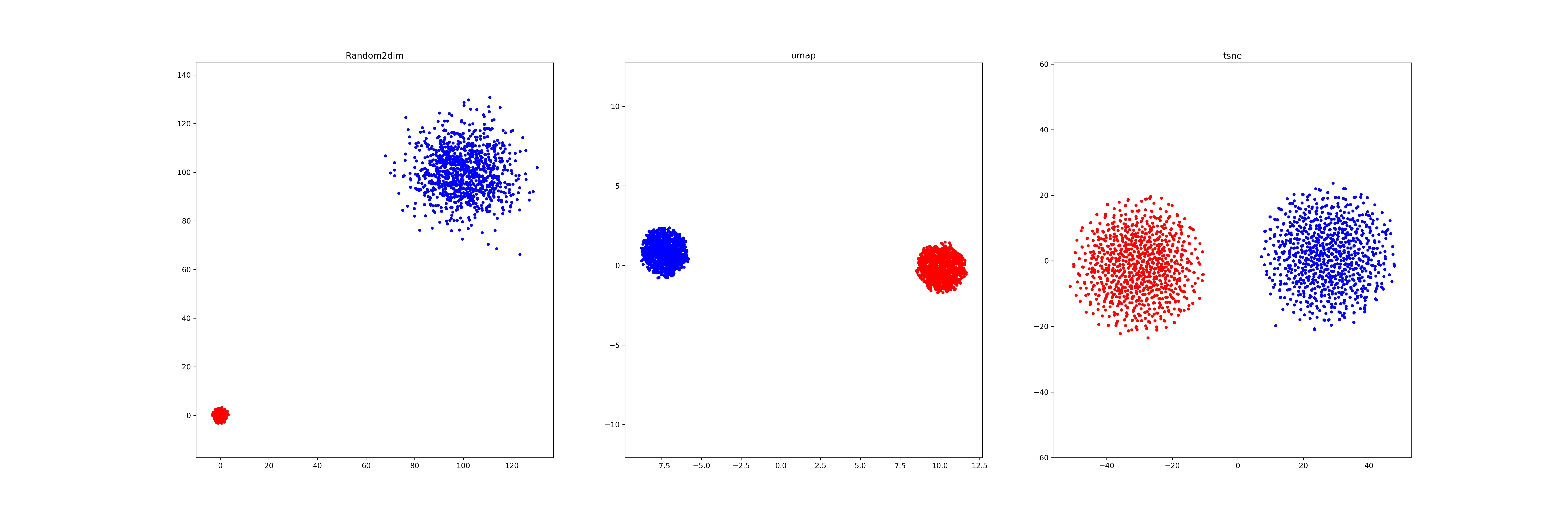
(2): We generate two different Gaussian distribution dataset (1000*64,1000*64) in different location(mean) and scale (std), and visualize it by a)random pick two dimension b) umap, c) tsne.
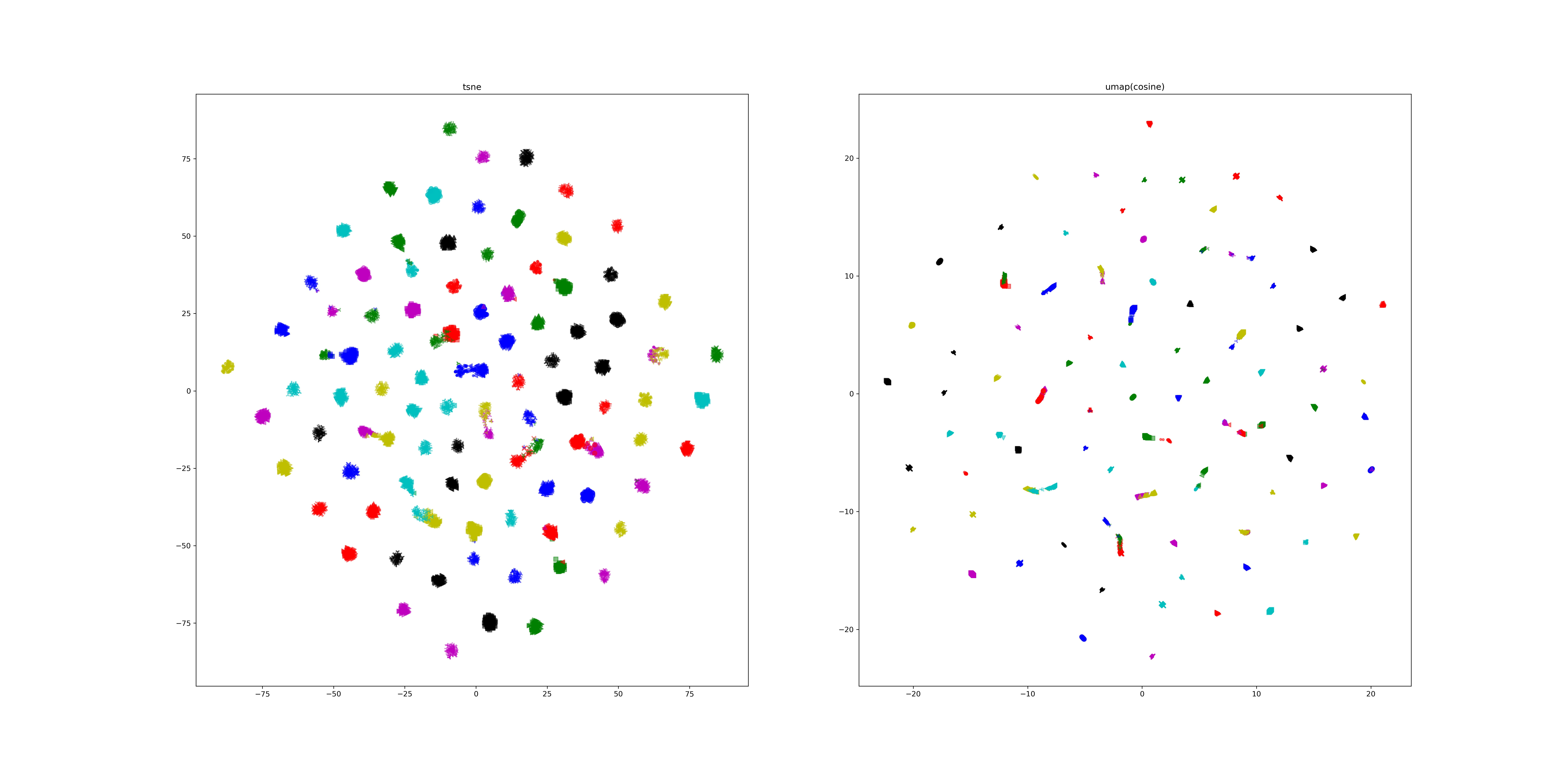
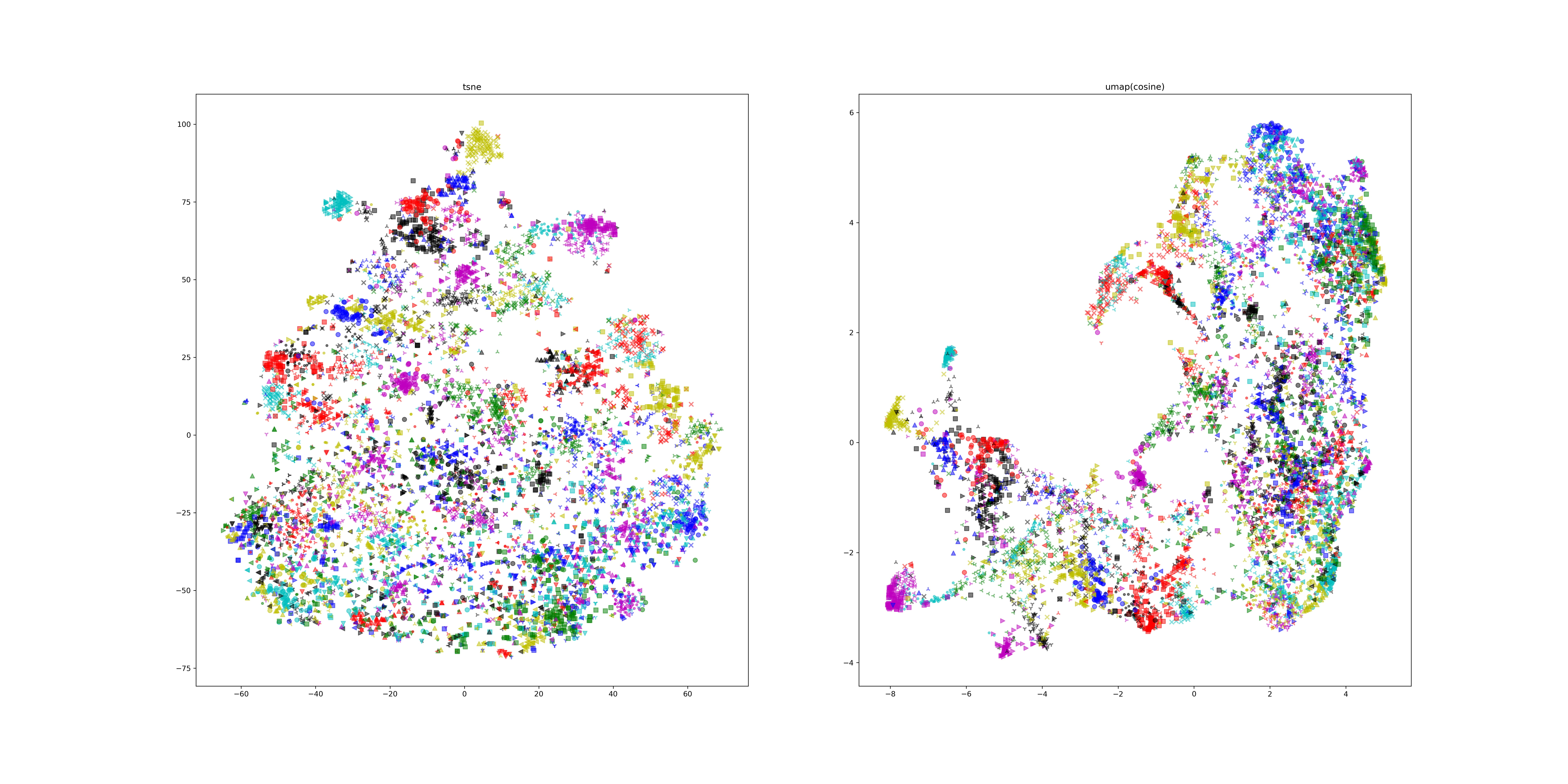
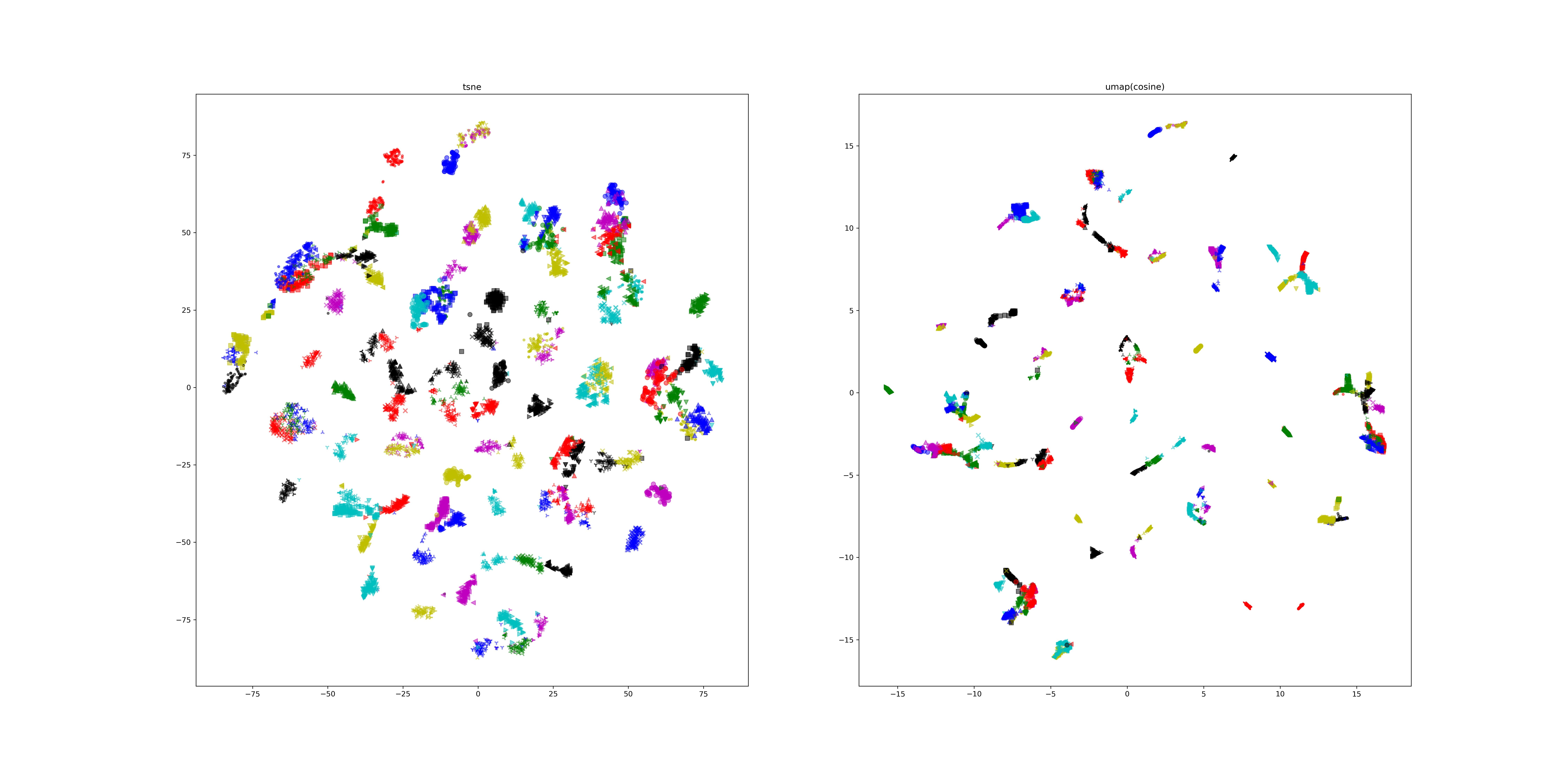
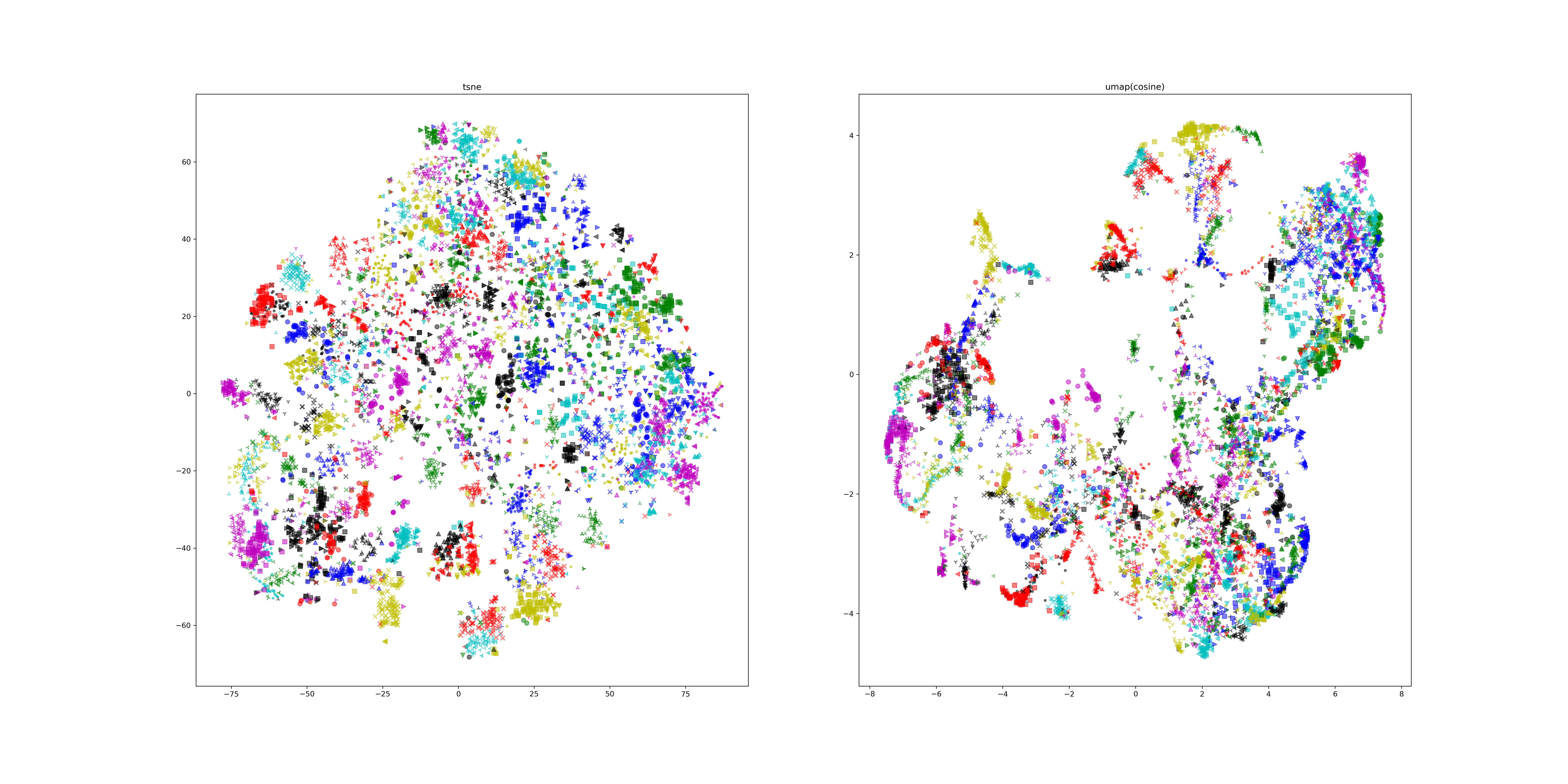
Then, we try to compare result of t-SNE and UMAP on embedding result of npair and epshn, on CAR dataset.
Npair on training data:
Npair val data:
EPSHN tra data:
EPSHN val data: