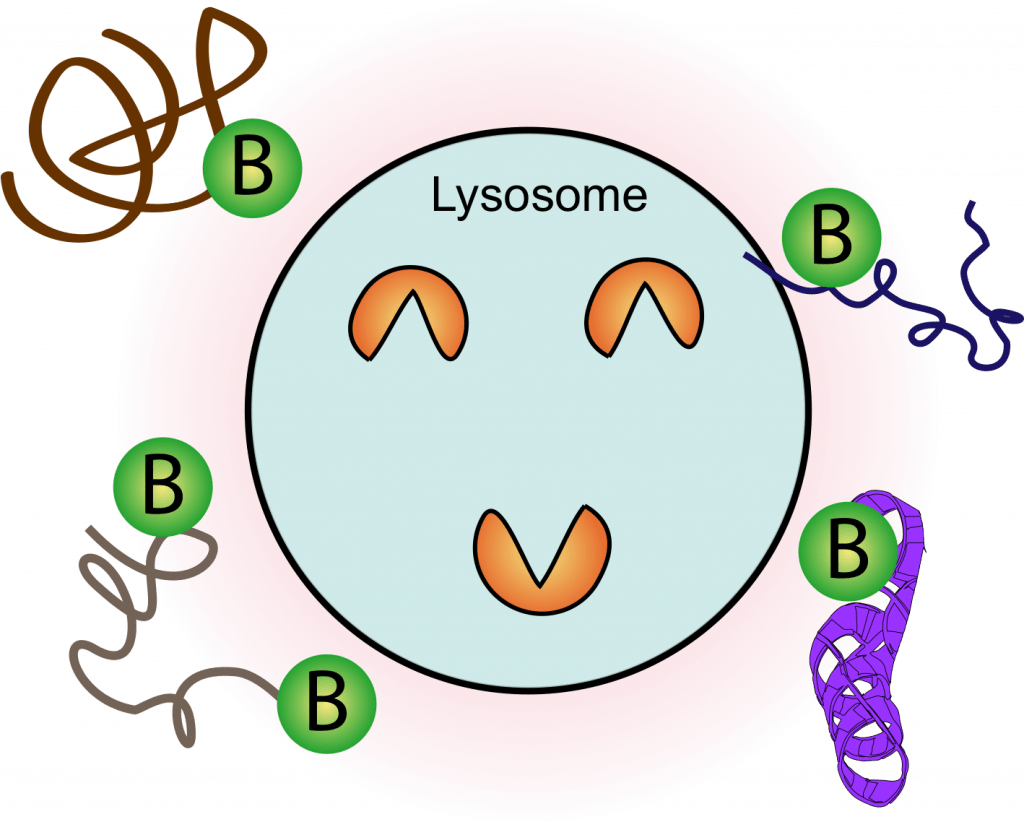
Proximity labeling MS of the Lysosome
Lysosomes are the terminal sites to break down and recycle cellular wastes through the autophagy pathway. We develop novel and improved proximity labeling mass spectrometry methods to study the highly dynamic microenvironment of the lysosomes in a high-throughput and quantitative fashion.

Dynamic Protein Turnover in Neurons
Dysregulated protein quality control is involved in aging and various diseases. We develop dynamic SILAC proteomic methods to quantify protein turnover rates in human iPSC-derived neurons and the changes of protein half-lives under genetic and cellular perturbations.
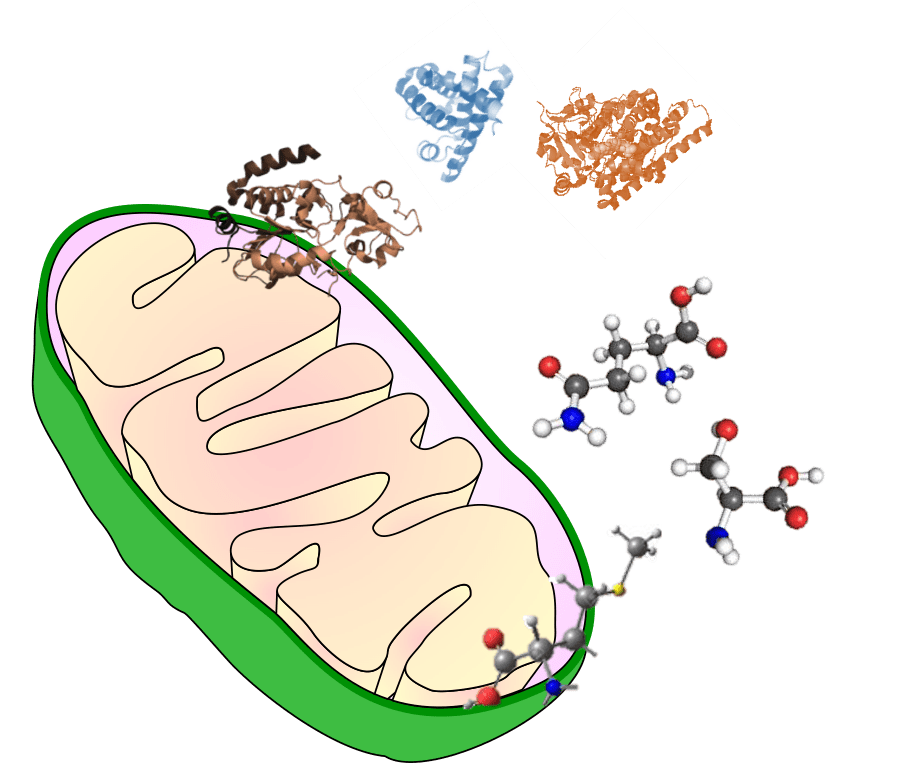
Mitochondrial Proteomics and Metabolomics
Mitochondria are the powerhouses and metabolic hubs of the cell. We develop untargeted and targeted LC-MS-based omics approaches to study the mitochondrial dysfunctions in neurodegenerative diseases.
More Collaborative Projects:
Multi-omics evaluation to study neurodevelopmental disease using patient fibroblasts.
(in collaboration with Prof. Anne Chiaramello’s group at GW School of Medicine)
3D cancer proteomics in breast cancer mouse model.
(in collaboration with Prof. Rong Li’s group at GW School of Medicine)
Protein interaction studies to understand fundamental mitochondrial biology.
(in collaboration with Prof. Shiori Sekine’s group at University of Pittsburgh)
