
Research Overview:
Quantitative analysis of biomolecules offers a wealth of clues to understand cell biology and disease mechanisms. Mass spectrometry (MS) has become a central technology to study biomolecules such as proteins, peptides, lipids, and small molecule metabolites. Our research focuses on developing novel and improved bioanalytical methods using LC-MS platforms and applying the combination of analytical chemistry, bioinformatics, and cell biology approaches to study human diseases, in particular, neurodegenerative diseases.
An interview video with Prof. Ling Hao: Chemistry’s Role in Studying Brain Diseases
Neurodegenerative Diseases
Neurodegeneration refers to the progressive loss of function/structure and eventually death of neurons in the brain, e.g. Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, Frontotemporal dementia, Amyotrophic lateral sclerosis, etc. Unfortunately, there is currently no cure for these diseases due to our limited understanding of the molecular mechanisms that provoke neurodegeneration. Through collaborative efforts with biologists and physician scientists, we aim to decipher the molecular mechanisms of neurodegeneration, in particular, lysosomal, mitochondrial, and neurotransmitter dysfunctions via human iPSC-derived neurons and mouse models of neurodegenerative diseases.
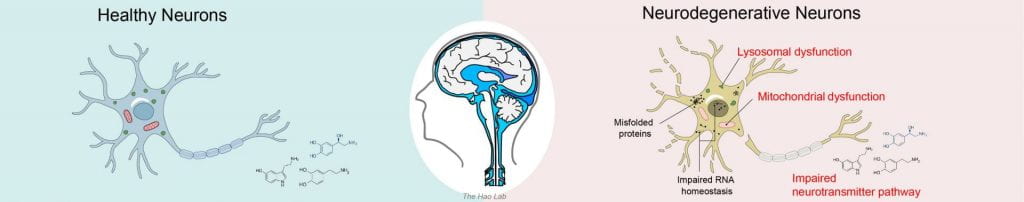
Human stem cell-derived neurons
In 2006, Human skin dermal fibroblasts was first reprogrammed into induced pluripotent stem cells (iPSCs) (2012 Nobel Prize). Human iPSC can be further differentiated into various cell types to model human diseases in vitro. Particularly, iPSC-derived neurons enable the study of live human neurons that were previously inaccessible, bridging the gap between animal models and human patient to study the human brain. Our lab cultures iPSC-derived neurons and conduct mass spectrometry-based omics on the i3Neuron platform to discover disease biomarkers and molecular mechanisms of neurodegenerative diseases.
MS-based Proteomics:
Proteins are the building blocks of life. Proteomics is the large-scale study of the complete set of proteins within a given biological system. With the rapid technological advancements, we can determine the compositions, dynamic changes, modifications, structures, distributions, interactions and half-lives of proteins via a variety of MS-based platforms. Our lab aims to develop new and improved LC-MS-based proteomic strategies (both untargeted and targeted methods) to understand the protein/peptide perturbations underlying human diseases.
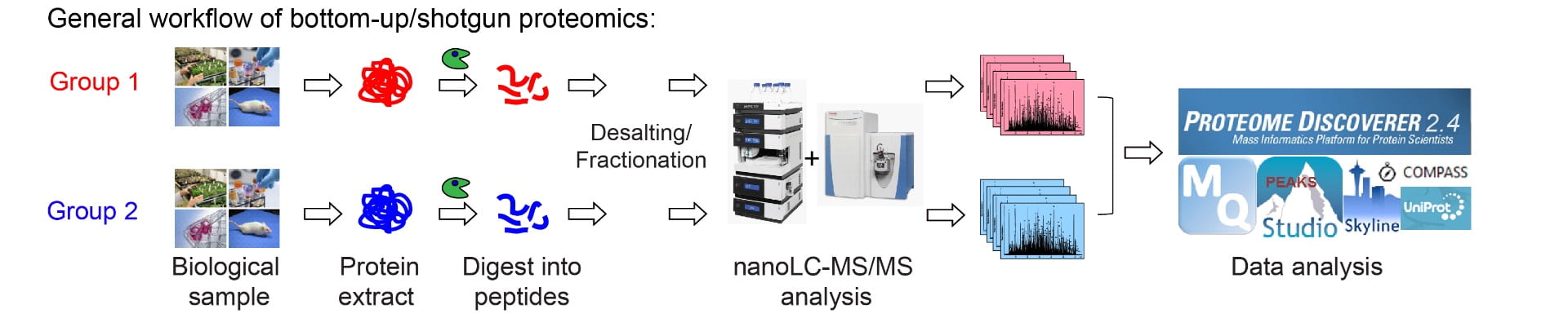
Proximity Labeling MS:
Proteins rarely function by themselves. Many cellular processes are dependent on protein-protein interactions. These interactions can be stable and permanent (e.g. structural protein complex), and they can also be weak and transient (e.g. signaling transduction). Affinity purification is typically used to capture the stable protein interactions. Proximity labeling (PL) techniques, developed in recent years, provide the capability to capture both stable and transient protein interactions as well as molecular microenvironment in living cells and organisms. Our lab aims to develop PL-MS strategies to achieve optimal sensitivity, specificity, and quantitative accuracy to study protein-protein/organelle interactions in human iPSC-derived neurons.
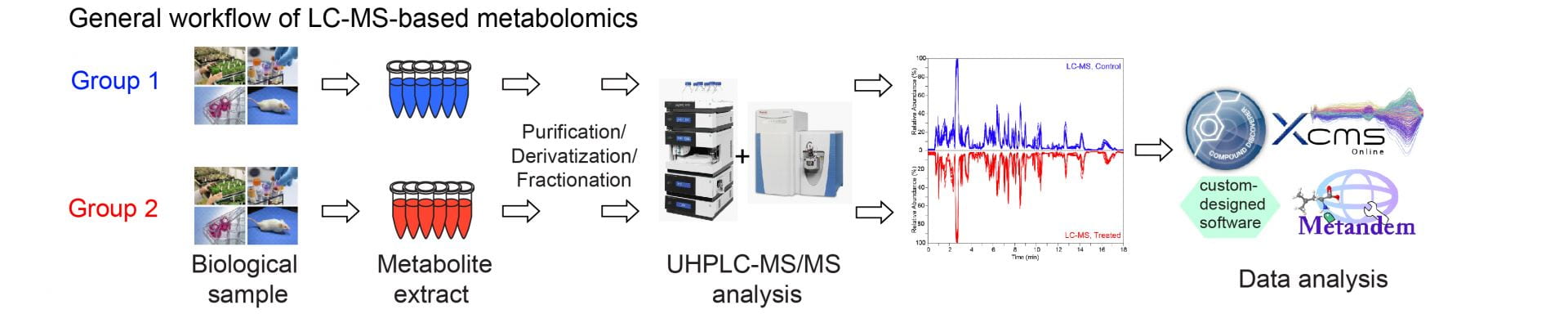
MS-based Metabolomics:
Small molecule metabolites provide direct functional readout of cellular processes and phenotypes. Metabolomics is the systematic study of small molecules within a given biological system. Global metabolomic profiling and targeted quantification have been widely used for disease biomarker discovery and clinical diagnosis. Our lab aims to develop MS-based untargeted and targeted metabolomic methods, bioinformatic tools, and multi-omic integrations to study the molecular pathways in human diseases.
— Proximity labeling proteomics to study the lysosomal microenvironment in human iPSC-derived neurons.
— Dynamic protein turnover in neuronal model of Frontotemporal dementia.
— Multi-omics characterization of mitochondrial dysfunctions in Parkinson’s disease.
Research Fundings:
- CAREER Award, National Science Foundation (NSF)
- Cottrell Scholar, Research Corporation For Science Advancement (RCSA)
- R01 grant, National Institutes of Health (NIH)
- UFF; CDRF, Office of the Vice President for Research, GW
- Ralph E. Powe Junior Faculty Enhancement Award, ORAU
- Faculty Startup Fund, GW
Relevant review literatures:
- Aebersold, R., and Matthias M. “Mass spectrometry-based proteomics.” Nature 422.6928 (2003): 198-207.
- Han, S., Li, J. and Ting, A.Y. “Proximity labeling: spatially resolved proteomic mapping for neurobiology.” Current opinion in neurobiology 50 (2018): 17-23.
- Johnson, C.H., Ivanisevic, J., and Siuzdak, G. “Metabolomics: beyond biomarkers and towards mechanisms.” Nature reviews Molecular cell biology 17.7 (2016): 451-459.
Relevant Websites:





